Evolution of non-coding regions
The Wnt genes constitute a gene family that encode for secreted glycoproteins, which act as key intercellular signaling molecules during animal development. Wnt proteins regulate processes as diverse as segmentation, patterning of tissues and organs, as well as the control of asymmetric cell division and cell movements in invertebrates and vertebrates (Wodarz and Nusse 1998; Annu. Rev. Cell Dev. Biol. 14: 59-88). The best characterised member of the Wnt family is the wingless (wg) gene, the Wnt-1 homologue of the fruit fly, Drosophila melanogaster. wg functions in multiple tissues and its accurate regulation is required to ensure normal development.
Although
the mechanisms of wg signaling
are beginning to be understood (Hlsken and Behrens 2000; J Cell Sci. 113: 3545-3546),
much less is known about how the expression of wg
is regulated. Its regulation is affected by several signaling pathways (dpp/TGF-β, hedgehog, Notch;
Neumann and Cohen 1997; Bioassays 19: 721-729)
and transcription factors like homothorax
(Casares and Mann 2000; Development 127: 1499-1508).
A larval enhancer (Neumann and Cohen 1996; Development 122: 1781-1789)
and an embryonic enhancer (Lessing and Nusse 1998; Development 125: 1469-1476)
have been identified within the 5’ wg
region. Nevertheless, these enhancers do not account for the whole
expression of the gene and, furthermore, there is very little information
on the cis-acting sequences and
regulatory molecules responsible for their regulatory activity. Therefore,
the study of how the accurate expression of wg
is achieved is critical for understanding the role of wg/Wnt in development and
disease.
An
important step towards the understanding of gene regulation is the
capability to identify regulatory elements associated with a given gene.
Most of these regulatory elements are relatively short stretches of DNA (5
to 25 nucleotide-long), located in the non-coding sequence surrounding a
gene. Most known transcription factor binding sites are located 5’ of
the coding region, but some are also found in the 3’ sequence, and even
in introns (in all these cases, regulatory elements are located embedded
in otherwise non-functional sequences). Because functional sequences are
maintained in evolution, they often can be recognized by their
conservation among different organisms. Thus, comparative DNA sequence
analysis is a powerful technique to identify regulatory elements. In the
only comparative study to date of Drosophila
wg non-coding sequences, Lessing
and Nusse (1998; Development 125: 1469-1476)
have compared 4.5 kb of the 5' non-coding flanking region of wg
from D. melanogaster and D.
virilis,
two species diverging for the last 40-60 My (Russo et al. 1995; Mol.
Biol. Evol. 12: 391-404).
This approach has led to the identification of 19 conserved sequence
blocks that are involved in the regulation of the expression of this gene.
Most of these elements are not repeated anywhere else in the D.
melanogaster genome (unpublished data), and could not therefore have
been easily identified without a comparative approach. This approach is
accompanied by population studies that can give further insight into the
relative strength of selection in different regions and sub-regions.
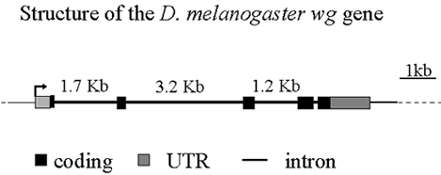
Together with Fernando Casares' Group, we are currently in the process of identifying regulatory elements in wg introns and 3' non-coding flanking regions and inferring the mode of evolution of wg regulatory elements.
![]()

Researchers involved in this project:
Collaborators:
Fernando Casares